Functional programming and iteration with purrr
2020-08-22
1 / 90
purrr: A functional programming toolkit for R
Complete and consistent set of tools for working with functions and vectors
2 / 90
Problems we want to solve:
- Making code clear
- Making code safe
- Working with lists and data frames
3 / 90
Lists, vectors, and data.frames (or tibbles)
c(char = "hello", num = 1)## char num ## "hello" "1"4 / 90
lists can contain any object
list(char = "hello", num = 1, fun = mean)## $char## [1] "hello"## ## $num## [1] 1## ## $fun## function (x, ...) ## UseMethod("mean")## <bytecode: 0x7fb922834d08>## <environment: namespace:base>5 / 90
Your Turn 1
measurements <- list( blood_glucose = rnorm(10, mean = 140, sd = 10), age = rnorm(5, mean = 40, sd = 5), heartrate = rnorm(20, mean = 80, sd = 15))There are two ways to subset lists: dollar signs and brackets. Try to subset blood_glucose from measurements using these approaches. Are they different? What about measurements[["blood_glucose"]]?
6 / 90
Your Turn 1
measurements["blood_glucose"]## $blood_glucose## [1] 127.9293 142.7743 150.8444 116.5430 144.2912 145.0606 134.2526 134.5337 134.3555 131.0996measurements$blood_glucose## [1] 127.9293 142.7743 150.8444 116.5430 144.2912 145.0606 134.2526 134.5337 134.3555 131.0996measurements[["blood_glucose"]]## [1] 127.9293 142.7743 150.8444 116.5430 144.2912 145.0606 134.2526 134.5337 134.3555 131.09967 / 90
data frames are lists
x <- list(char = "hello", num = 1)as.data.frame(x)## char num## 1 hello 18 / 90
data frames are lists
library(gapminder)head(gapminder$pop)## [1] 8425333 9240934 10267083 11537966 13079460 148803729 / 90
data frames are lists
gapminder[1:6, "pop"]10 / 90
data frames are lists
gapminder[1:6, "pop"]## # A tibble: 6 x 1## pop## <int>## 1 8425333## 2 9240934## 3 10267083## 4 11537966## 5 13079460## 6 1488037211 / 90
data frames are lists
head(gapminder[["pop"]])## [1] 8425333 9240934 10267083 11537966 13079460 1488037212 / 90
vectorized functions don't work on lists
sum(rnorm(10))13 / 90
vectorized functions don't work on lists
sum(rnorm(10))## [1] -3.83157414 / 90
vectorized functions don't work on lists
sum(rnorm(10))## [1] -3.831574sum(list(x = rnorm(10), y = rnorm(10), z = rnorm(10)))15 / 90
vectorized functions don't work on lists
sum(rnorm(10))## [1] -3.831574sum(list(x = rnorm(10), y = rnorm(10), z = rnorm(10)))## Error in sum(list(x = rnorm(10), y = rnorm(10), z = rnorm(10))): invalid 'type' (list) of argument16 / 90
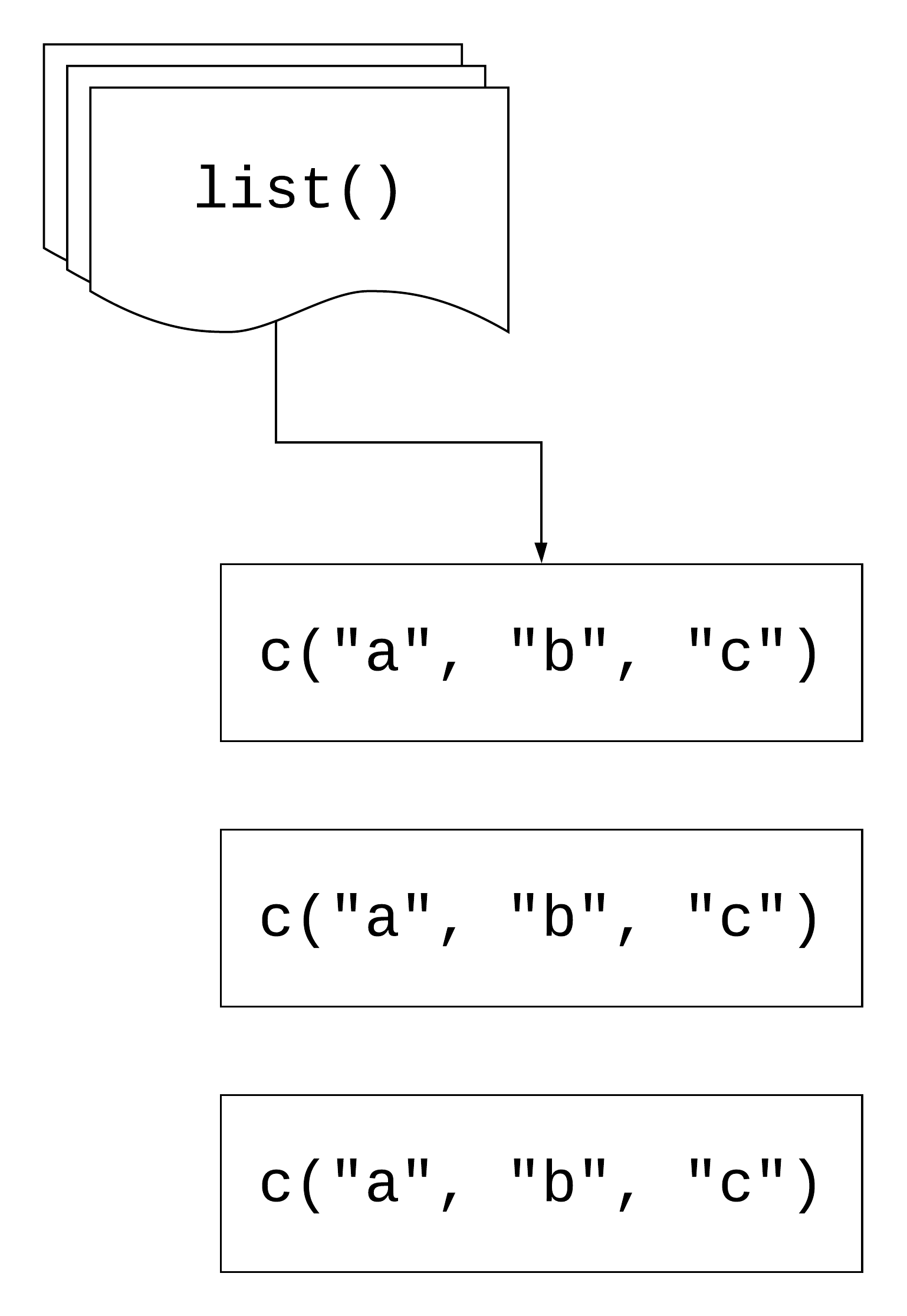
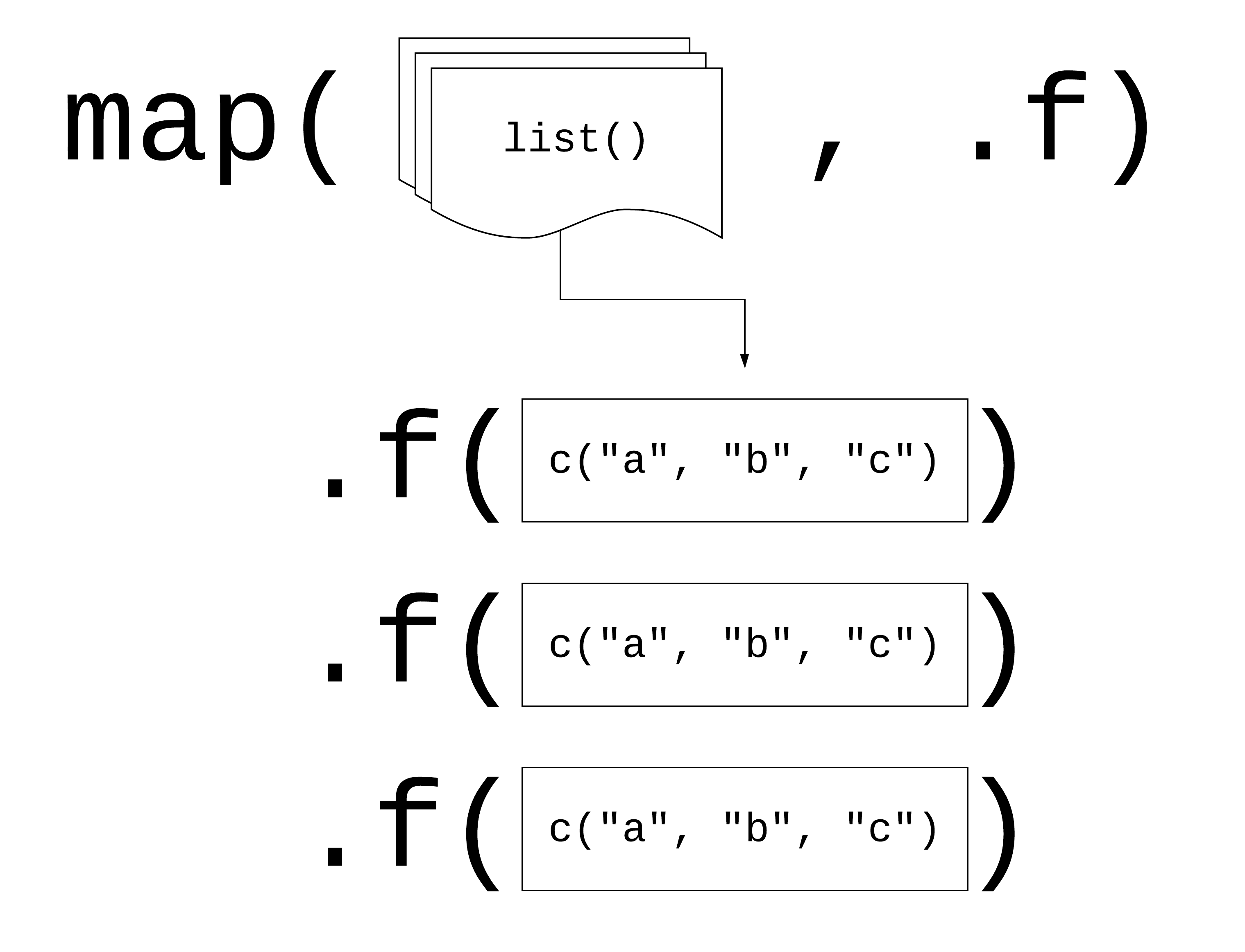
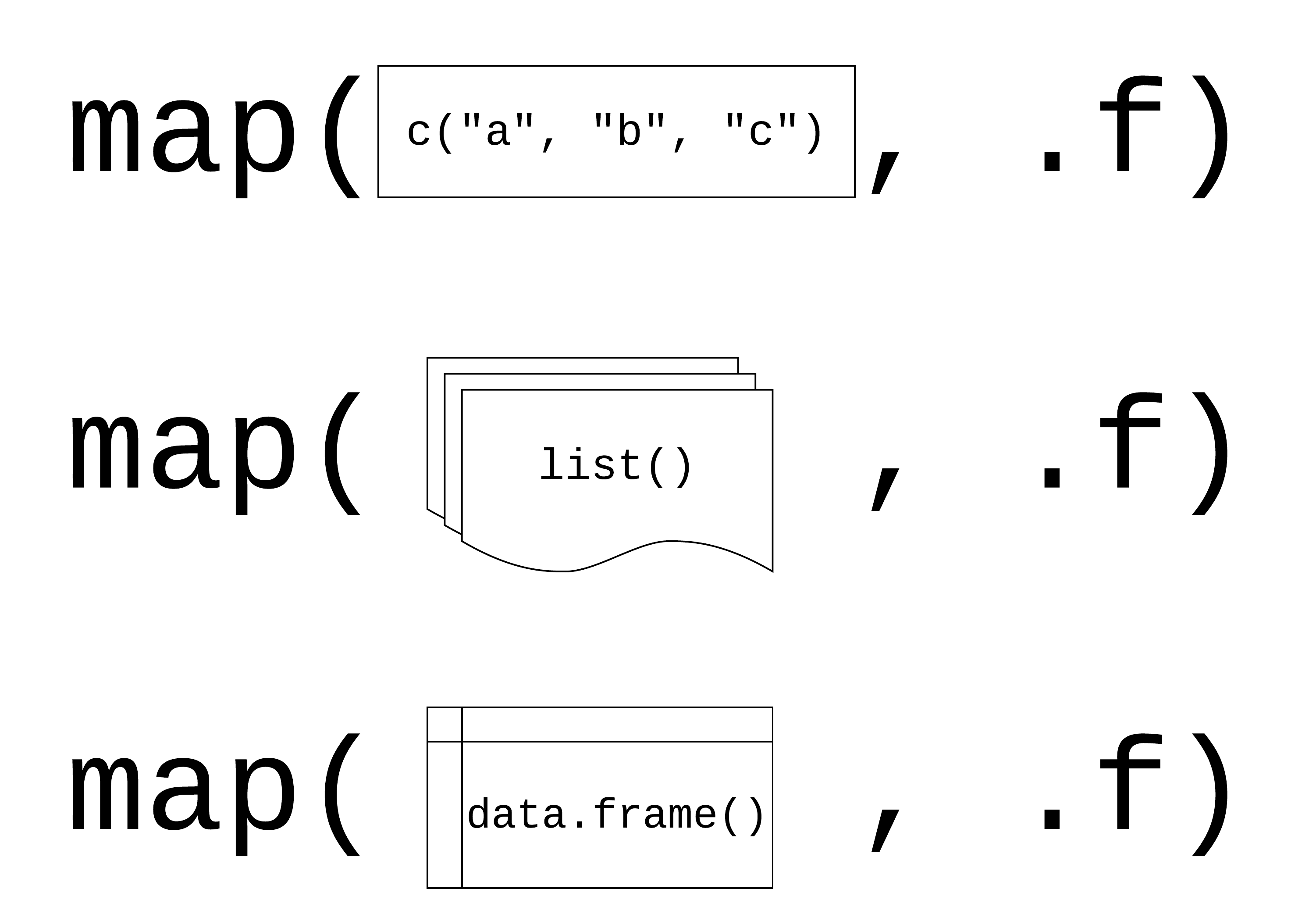
map(.x, .f)
17 / 90
map(.x, .f)
.x: a vector, list, or data frame
17 / 90
map(.x, .f)
.x: a vector, list, or data frame
.f: a function
17 / 90
map(.x, .f)
.x: a vector, list, or data frame
.f: a function
Returns a list
17 / 90
Using map()
library(purrr)x_list <- list(x = rnorm(10), y = rnorm(10), z = rnorm(10))map(x_list, mean)18 / 90
Using map()
library(purrr)x_list <- list(x = rnorm(10), y = rnorm(10), z = rnorm(10))map(x_list, mean)19 / 90
Using map()
library(purrr)x_list <- list(x = rnorm(10), y = rnorm(10), z = rnorm(10))map(x_list, mean)20 / 90
Using map()
library(purrr)x_list <- list(x = rnorm(10), y = rnorm(10), z = rnorm(10))map(x_list, mean)## $x## [1] -0.6097971## ## $y## [1] -0.2788647## ## $z## [1] 0.616592221 / 90
22 / 90
23 / 90
24 / 90
Your Turn 2
Read the code in the first chunk and predict what will happen
Run the code in the first chunk. What does it return?
list( sum_blood_glucose = sum(measurements$blood_glucose), sum_age = sum(measurements$age), sum_heartrate = sum(measurements$heartrate))Now, use map() to create the same output.
25 / 90
Your Turn 2
map(measurements, sum)## $blood_glucose## [1] 1361.684## ## $age## [1] 193.8606## ## $heartrate## [1] 1509.30426 / 90
using map() with data frames
27 / 90
using map() with data frames
library(dplyr)gapminder %>% select(where(is.numeric)) %>% map(sd)27 / 90
using map() with data frames
library(dplyr)gapminder %>% select(where(is.numeric)) %>% map(sd)28 / 90
using map() with data frames
library(dplyr)gapminder %>% select(where(is.numeric)) %>% map(sd)29 / 90
using map() with data frames
library(dplyr)gapminder %>% select(where(is.numeric)) %>% map(sd)## $year## [1] 17.26533## ## $lifeExp## [1] 12.91711## ## $pop## [1] 106157897## ## $gdpPercap## [1] 9857.45530 / 90
Your Turn 3
Pass diabetes to map() and map using class(). What are these results telling you?
31 / 90
Your Turn 3
head( map(diabetes, class), 3)## $id## [1] "numeric"## ## $chol## [1] "numeric"## ## $stab.glu## [1] "numeric"32 / 90
Review: writing functions
x <- x^2x <- scale(x)x <- max(x)33 / 90
Review: writing functions
x <- x^2x <- scale(x)x <- max(x)y <- x^2y <- scale(y)y <- max(y)z <- z^2z <- scale(x)z <- max(z)34 / 90
Review: writing functions
x <- x^2x <- scale(x)x <- max(x)y <- x^2y <- scale(y)y <- max(y)z <- z^2z <- scale(x)z <- max(z)35 / 90
Review: writing functions
x <- x^3x <- scale(x)x <- max(x)y <- x^2y <- scale(y)y <- max(y)z <- z^2z <- scale(x)z <- max(z)36 / 90
Review: writing functions
.f <- function(x) { x <- x^3 x <- scale(x) max(x)}.f(x).f(y).f(z)37 / 90
If you copy and paste your code three times, it's time to write a function
38 / 90
Your Turn 4
Write a function that returns the mean and standard deviation of a numeric vector.
Give the function a name
Find the mean and SD of x
Map your function to measurements
39 / 90
Your Turn 4
mean_sd <- function(x) { x_mean <- mean(x) x_sd <- sd(x) tibble(mean = x_mean, sd = x_sd)}map(measurements, mean_sd)40 / 90
Your Turn 4
## $blood_glucose## # A tibble: 1 x 2## mean sd## <dbl> <dbl>## 1 136. 9.96## ## $age## # A tibble: 1 x 2## mean sd## <dbl> <dbl>## 1 38.8 3.91## ## $heartrate## # A tibble: 1 x 2## mean sd## <dbl> <dbl>## 1 75.5 13.841 / 90
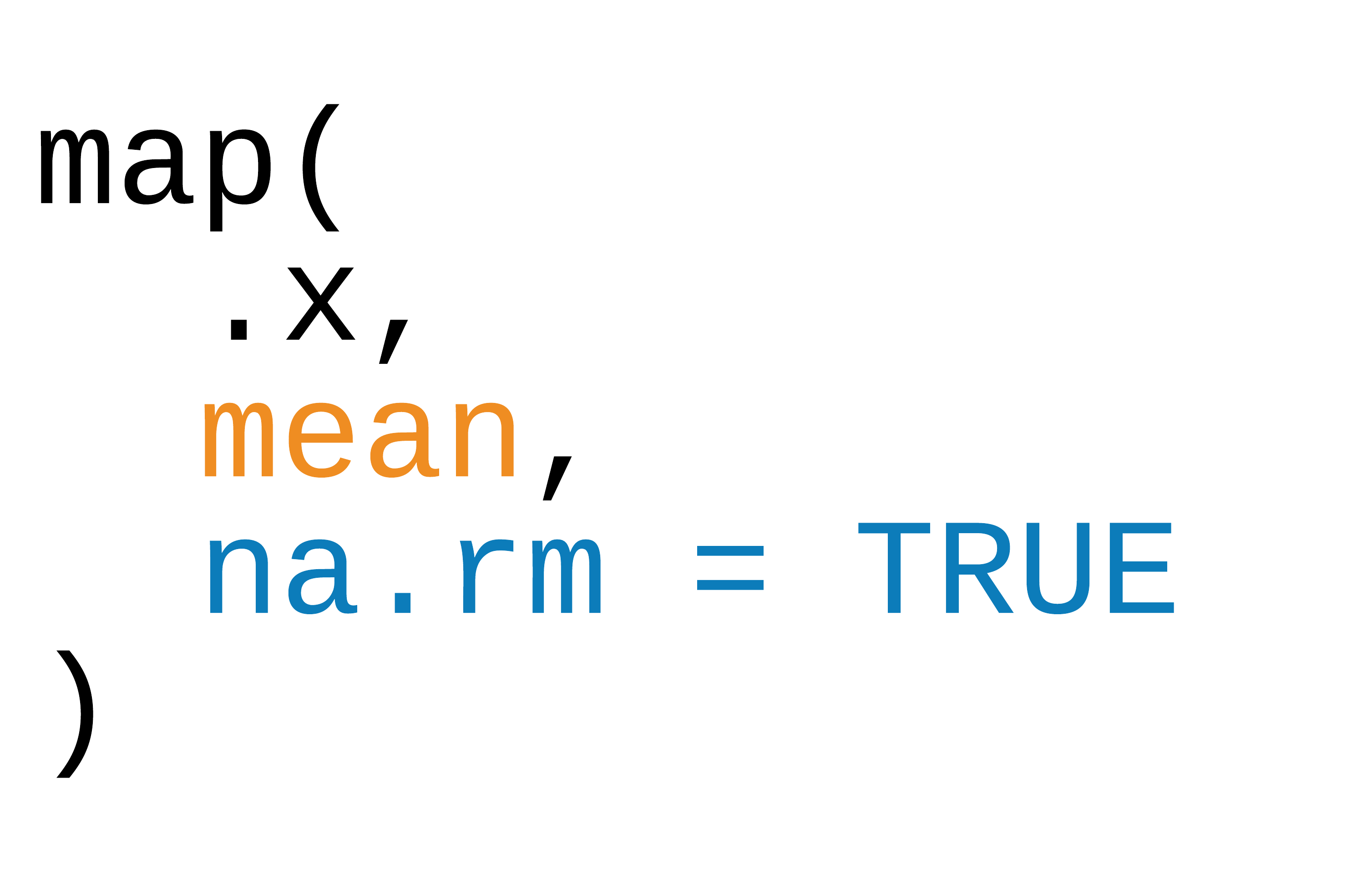
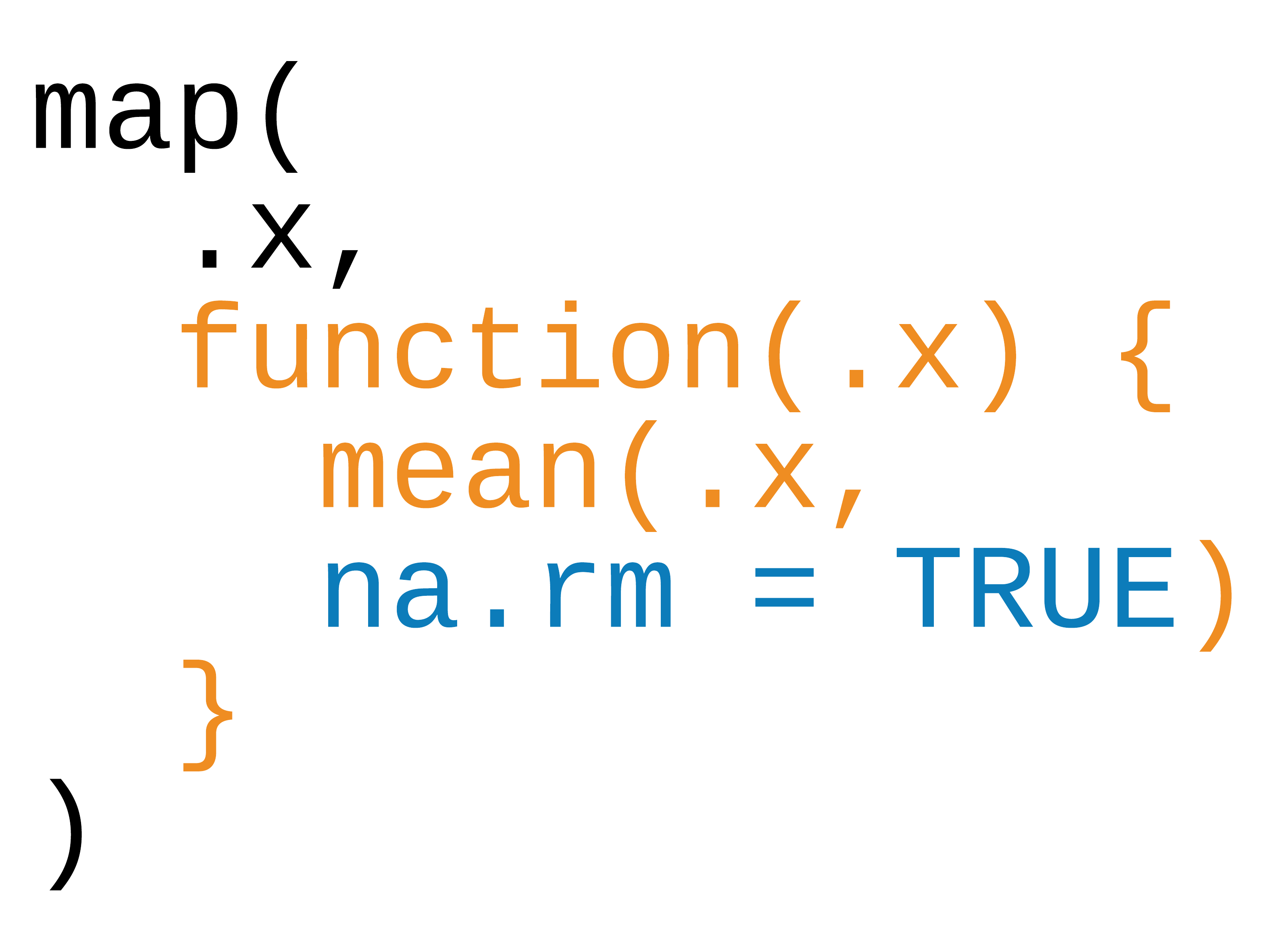
Three ways to pass functions to map()
- pass directly to
map() - use an anonymous function
- use ~
42 / 90
43 / 90
44 / 90
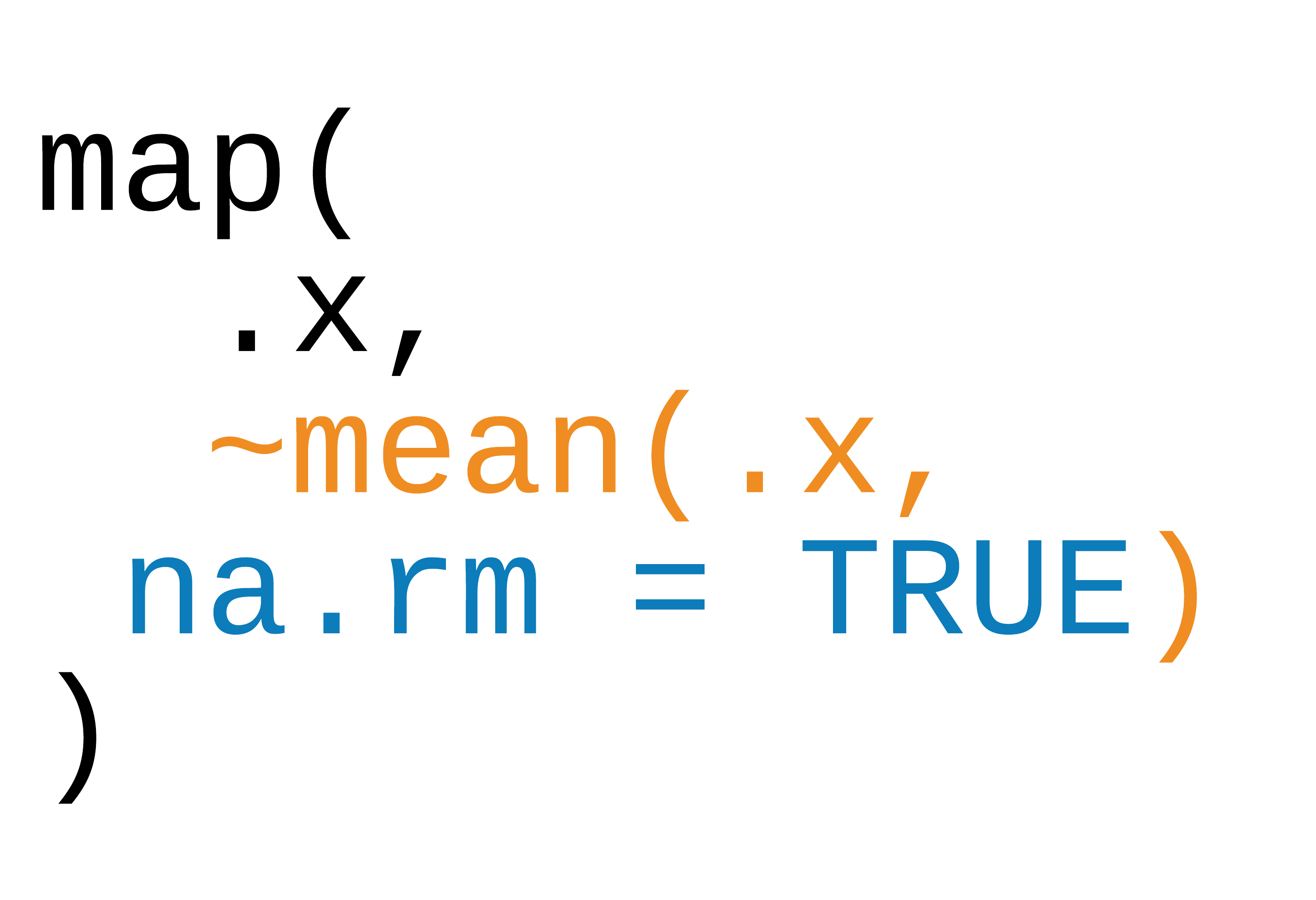
45 / 90
map(gapminder, ~length(unique(.x)))46 / 90
map(gapminder, ~length(unique(.x)))## $country## [1] 142## ## $continent## [1] 5## ## $year## [1] 12## ## $lifeExp## [1] 1626## ## $pop## [1] 1704## ## $gdpPercap## [1] 170447 / 90
Returning types
| map | returns |
|---|---|
map() |
list |
map_chr() |
character vector |
map_dbl() |
double vector (numeric) |
map_int() |
integer vector |
map_lgl() |
logical vector |
map_dfc() |
data frame (by column) |
map_dfr() |
data frame (by row) |
48 / 90
Returning types
map_int(gapminder, ~length(unique(.x)))49 / 90
Returning types
map_int(gapminder, ~length(unique(.x)))## country continent year lifeExp pop gdpPercap ## 142 5 12 1626 1704 170450 / 90
Your Turn 5
Do the same as #3 above but return a vector instead of a list.
51 / 90
Your Turn 5
map_chr(diabetes, class)## id chol stab.glu hdl ratio glyhb location age gender height weight frame bp.1s bp.1d bp.2s ## "numeric" "numeric" "numeric" "numeric" "numeric" "numeric" "character" "numeric" "character" "numeric" "numeric" "character" "numeric" "numeric" "numeric" ## bp.2d waist hip time.ppn ## "numeric" "numeric" "numeric" "numeric"52 / 90
Your Turn 6
Check diabetes for any missing data.
Using the ~.f(.x) shorthand, check each column for any missing values using is.na() and any()
Return a logical vector. Are any columns missing data? What happens if you don't include any()? Why?
Try counting the number of missing, returning an integer vector
53 / 90
Your Turn 6
map_lgl(diabetes, ~any(is.na(.x)))## id chol stab.glu hdl ratio glyhb location age gender height weight frame bp.1s bp.1d bp.2s bp.2d waist hip time.ppn ## FALSE TRUE FALSE TRUE TRUE TRUE FALSE FALSE FALSE TRUE TRUE TRUE TRUE TRUE TRUE TRUE TRUE TRUE TRUE54 / 90
Your Turn 6
map_int(diabetes, ~sum(is.na(.x)))## id chol stab.glu hdl ratio glyhb location age gender height weight frame bp.1s bp.1d bp.2s bp.2d waist hip time.ppn ## 0 1 0 1 1 13 0 0 0 5 1 12 5 5 262 262 2 2 355 / 90
Your Turn 7
Turn diabetes into a list split by location using the split() function. Check its length.
Fill in the model_lm function to model chol (the outcome) with ratio and pass the .data argument to lm()
map model_lm to diabetes_list so that it returns a data frame (by row).
56 / 90
Your Turn 7
diabetes_list <- split(diabetes, diabetes$location)length(diabetes_list)model_lm <- function(.data) { mdl <- lm(chol ~ ratio, data = .data) # get model statistics broom::glance(mdl)}map(diabetes_list, model_lm)57 / 90
Your Turn 7
## [1] 2## $Buckingham## # A tibble: 1 x 12## r.squared adj.r.squared sigma statistic p.value df## <dbl> <dbl> <dbl> <dbl> <dbl> <dbl>## 1 0.252 0.248 38.8 66.4 4.11e-14 1## # … with 6 more variables: logLik <dbl>, AIC <dbl>,## # BIC <dbl>, deviance <dbl>, df.residual <int>,## # nobs <int>## ## $Louisa## # A tibble: 1 x 12## r.squared adj.r.squared sigma statistic p.value df## <dbl> <dbl> <dbl> <dbl> <dbl> <dbl>## 1 0.204 0.201 39.4 51.7 1.26e-11 1## # … with 6 more variables: logLik <dbl>, AIC <dbl>,## # BIC <dbl>, deviance <dbl>, df.residual <int>,## # nobs <int>58 / 90
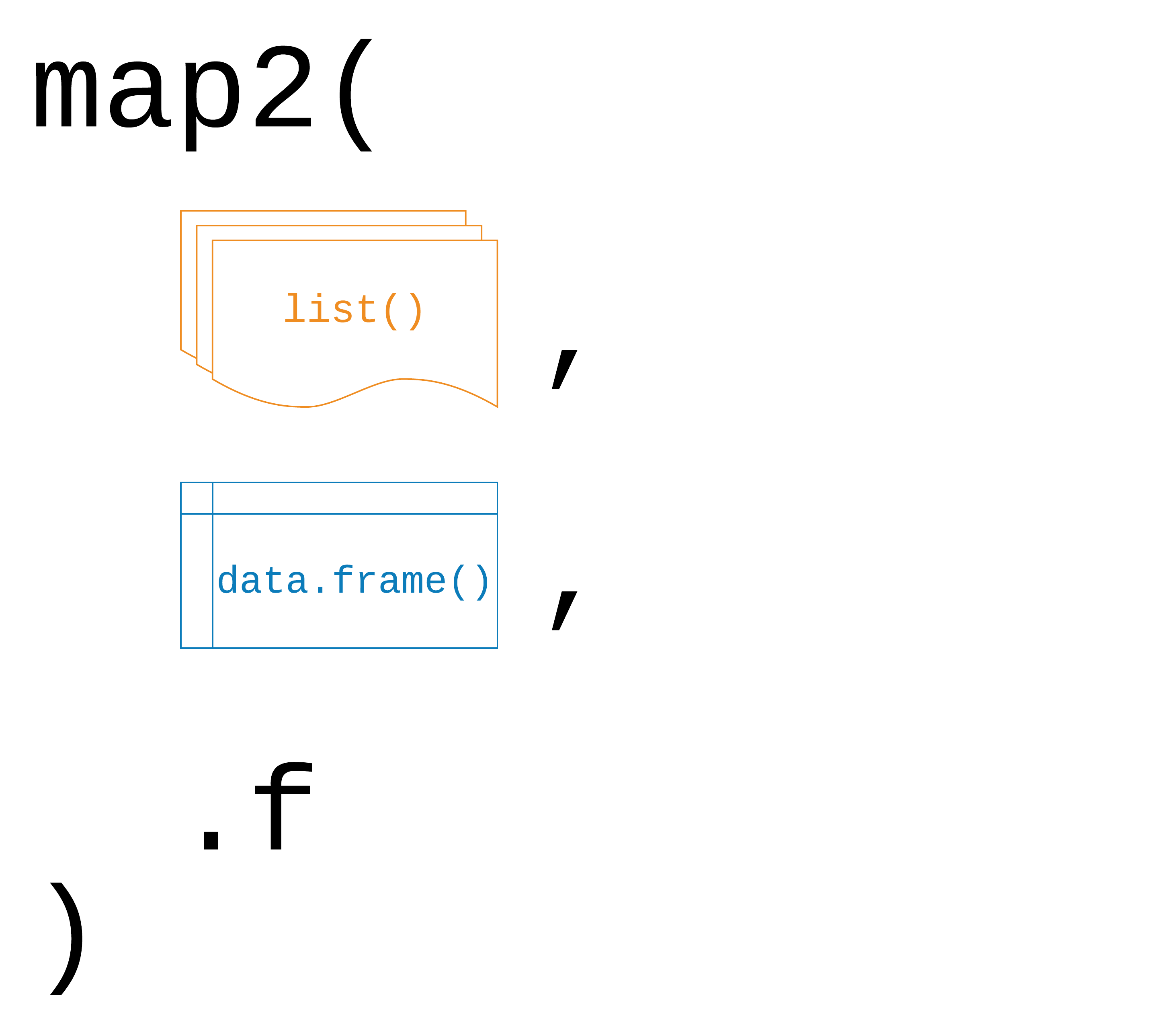
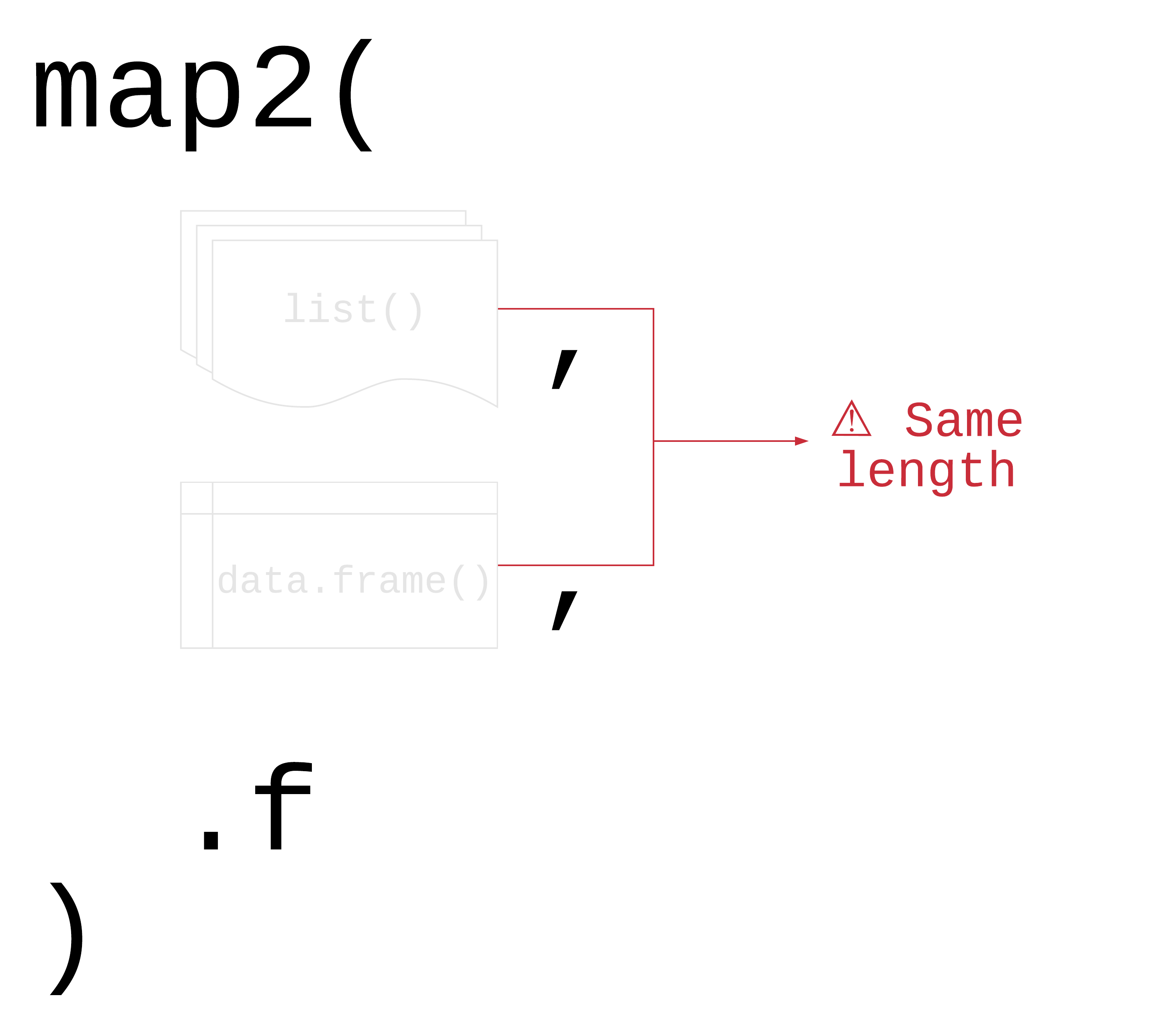
map2(.x, .y, .f)
59 / 90
map2(.x, .y, .f)
.x, .y: a vector, list, or data frame
59 / 90
map2(.x, .y, .f)
.x, .y: a vector, list, or data frame
.f: a function
59 / 90
map2(.x, .y, .f)
.x, .y: a vector, list, or data frame
.f: a function
Returns a list
59 / 90
60 / 90
61 / 90
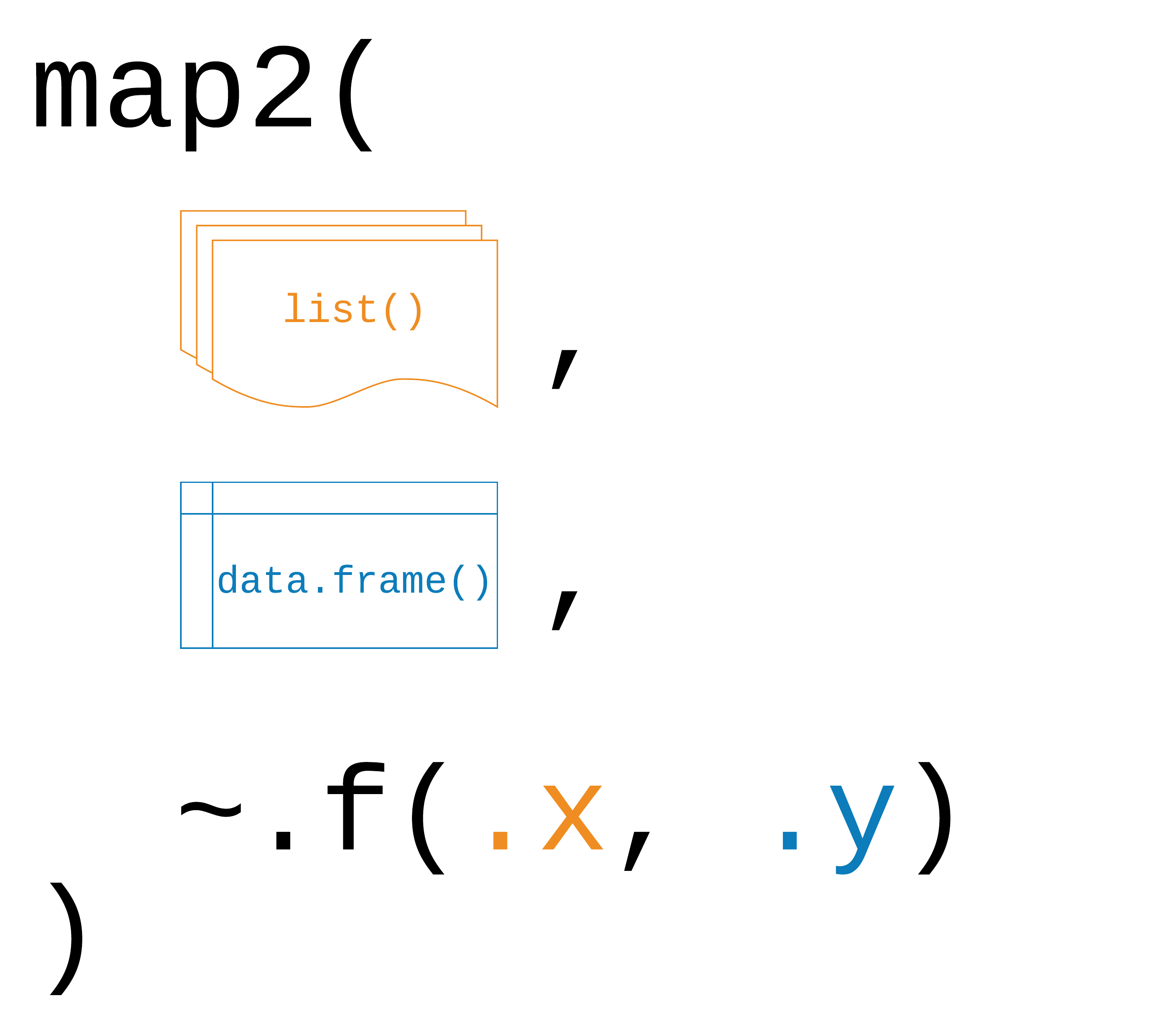
62 / 90
map2()
means <- c(-3, 4, 2, 2.3)sds <- c(.3, 4, 2, 1)map2_dbl(means, sds, rnorm, n = 1)63 / 90
map2()
means <- c(-3, 4, 2, 2.3)sds <- c(.3, 4, 2, 1)map2_dbl(means, sds, rnorm, n = 1)64 / 90
map2()
means <- c(-3, 4, 2, 2.3)sds <- c(.3, 4, 2, 1)map2_dbl(means, sds, rnorm, n = 1)## [1] -2.997932 2.178125 1.266952 2.94828765 / 90
Your Turn 8
Split the gapminder dataset into a list by country
Create a list of models using map(). For the first argument, pass gapminder_countries. For the second, use the ~.f() notation to write a model with lm(). Use lifeExp on the left hand side of the formula and year on the second. Pass .x to the data argument.
Use map2() to take the models list and the data set list and map them to predict(). Since we're not adding new arguments, you don't need to use ~.f().
66 / 90
Your Turn 8
gapminder_countries <- split(gapminder, gapminder$country)models <- map(gapminder_countries, ~ lm(lifeExp ~ year, data = .x))preds <- map2(models, gapminder_countries, predict)head(preds, 3)67 / 90
Your Turn 8
gapminder_countries <- split(gapminder, gapminder$country)models <- map(gapminder_countries, ~ lm(lifeExp ~ year, data = .x))preds <- map2(models, gapminder_countries, predict)head(preds, 3)68 / 90
Your Turn 8
gapminder_countries <- split(gapminder, gapminder$country)models <- map(gapminder_countries, ~ lm(lifeExp ~ year, data = .x))preds <- map2(models, gapminder_countries, predict)head(preds, 3)69 / 90
Your Turn 8
## $Afghanistan## 1 2 3 4 5 6 ## 29.90729 31.28394 32.66058 34.03722 35.41387 36.79051 ## ## $Albania## 1 2 3 4 5 6 ## 59.22913 60.90254 62.57596 64.24938 65.92279 67.59621 ## ## $Algeria## 1 2 3 4 5 6 ## 43.37497 46.22137 49.06777 51.91417 54.76057 57.6069770 / 90
| input 1 | input 2 | returns |
|---|---|---|
map() |
map2() |
list |
map_chr() |
map2_chr() |
character vector |
map_dbl() |
map2_dbl() |
double vector (numeric) |
map_int() |
map2_int() |
integer vector |
map_lgl() |
map2_lgl() |
logical vector |
map_dfc() |
map2_dfc() |
data frame (by column) |
map_dfr() |
map2_dfr() |
data frame (by row) |
71 / 90
Other mapping functions
pmap() and friends: take n lists or data frame with argument names
72 / 90
Other mapping functions
pmap() and friends: take n lists or data frame with argument names
walk() and friends: for side effects like plotting; returns input invisibly
73 / 90
Other mapping functions
pmap() and friends: take n lists or data frame with argument names
walk() and friends: for side effects like plotting; returns input invisibly
imap() and friends: includes counter i
74 / 90
Other mapping functions
pmap() and friends: take n lists or data frame with argument names
walk() and friends: for side effects like plotting; returns input invisibly
imap() and friends: includes counter i
imap_if(), map_at(): Apply only to certain elements
75 / 90
| input 1 | input 2 | input n | returns |
|---|---|---|---|
map() |
map2() |
pmap() |
list |
map_chr() |
map2_chr() |
pmap_chr() |
character vector |
map_dbl() |
map2_dbl() |
pmap_dbl() |
double vector (numeric) |
map_int() |
map2_int() |
pmap_int() |
integer vector |
map_lgl() |
map2_lgl() |
pmap_lgl() |
logical vector |
map_dfc() |
map2_dfc() |
pmap_dfc() |
data frame (by column) |
map_dfr() |
map2_dfr() |
pmap_dfr() |
data frame (by row) |
walk() |
walk2() |
pwalk() |
input (side effects!) |
76 / 90
Your turn 9
Create a new directory using the fs package. Call it "figures".
Write a function to plot a line plot of a given variable in gapminder over time, faceted by continent. Then, save the plot (how do you save a ggplot?). For the file name, paste together the folder, name of the variable, and extension so it follows the pattern "folder/variable_name.png"
Create a character vector that has the three variables we'll plot: "lifeExp", "pop", and "gdpPercap".
Use walk() to save a plot for each of the variables
77 / 90
Your turn 9
fs::dir_create("figures")ggsave_gapminder <- function(variable) { # we're using `aes_string()` so we don't need the curly-curly syntax p <- ggplot( gapminder, aes_string(x = "year", y = variable, color = "country") ) + geom_line() + scale_color_manual(values = country_colors) + facet_wrap(vars(continent.)) + theme(legend.position = "none") ggsave( filename = paste0("figures/", variable, ".png"), plot = p, dpi = 320 )}78 / 90
Your turn 9
vars <- c("lifeExp", "pop", "gdpPercap")walk(vars, ggsave_gapminder)79 / 90
Base R
| base R | purrr |
|---|---|
lapply() |
map() |
vapply() |
map_*() |
sapply() |
? |
x[] <- lapply() |
map_dfc() |
mapply() |
map2(), pmap() |
80 / 90
Benefits of purrr
- Consistent
- Type-safe
- ~f(.x)
81 / 90
Loops vs functional programming
x <- rnorm(10)y <- map(x, mean)x <- rnorm(10)y <- vector("list", length(x))for (i in seq_along(x)) { y[[i]] <- mean(x[[i]])}82 / 90
Loops vs functional programming
x <- rnorm(10)y <- map(x, mean)x <- rnorm(10) y <- vector("list", length(x))for (i in seq_along(x)) { y[[i]] <- mean(x[[i]]) }83 / 90
Loops vs functional programming
x <- rnorm(10)y <- map(x, mean)x <- rnorm(10)y <- vector("list", length(x)) for (i in seq_along(x)) { y[[i]] <- mean(x[[i]]) }84 / 90
Loops vs functional programming
x <- rnorm(10)y <- map(x, mean)x <- rnorm(10)y <- vector("list", length(x)) for (i in seq_along(x)) { y[[i]] <- mean(x[[i]])}85 / 90
Of course someone has to write loops. It doesn’t have to be you.
—Jenny Bryan
86 / 90
Working with lists and nested data
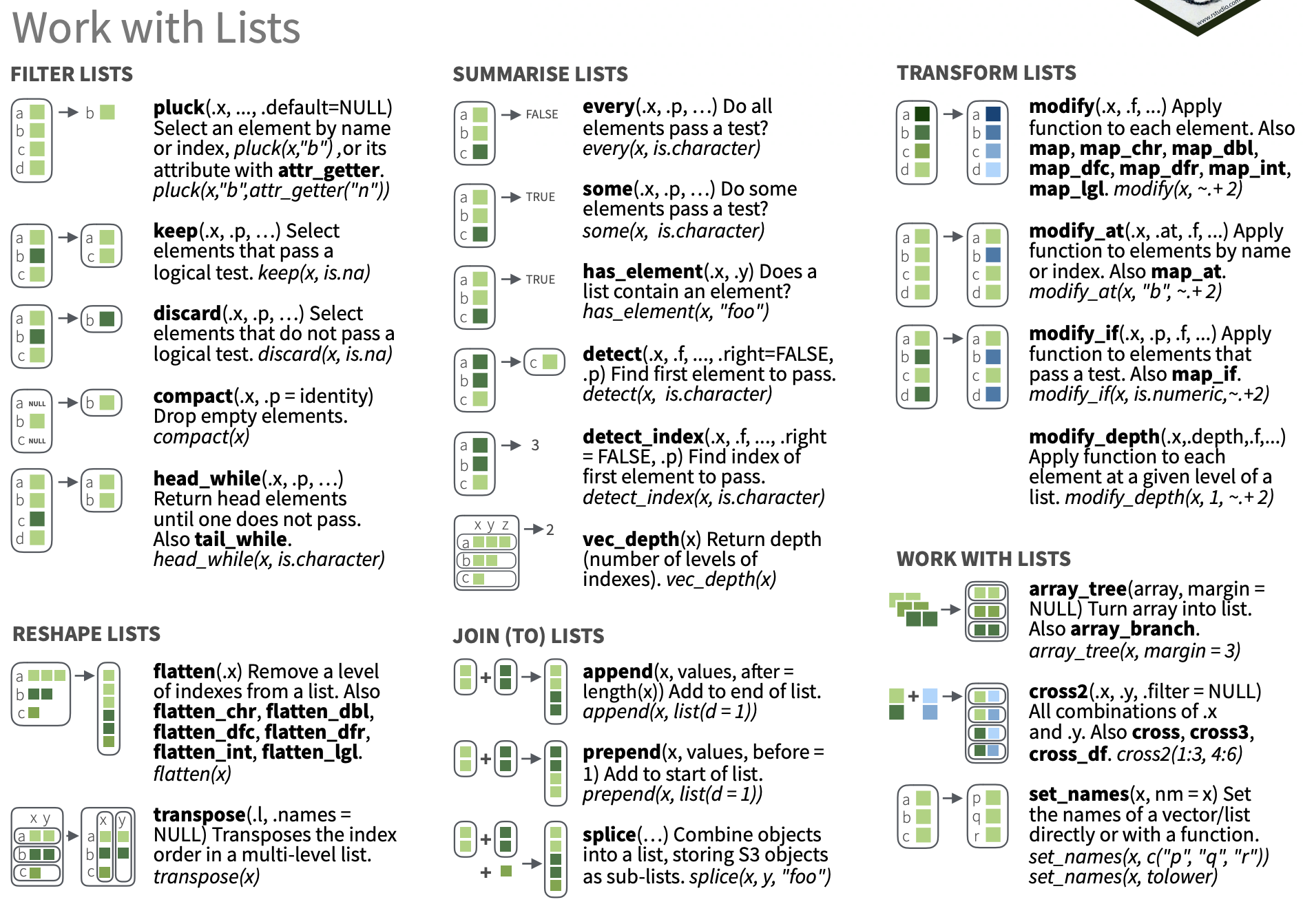
87 / 90
Working with lists and nested data
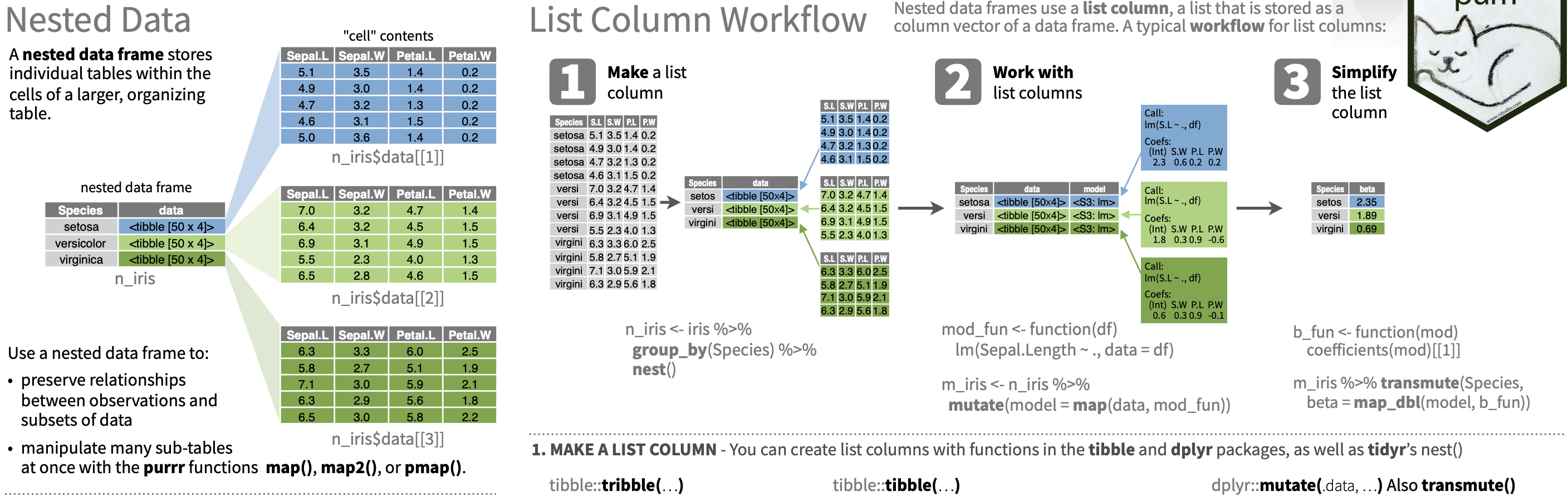
88 / 90
Adverbs: Modify function behavior
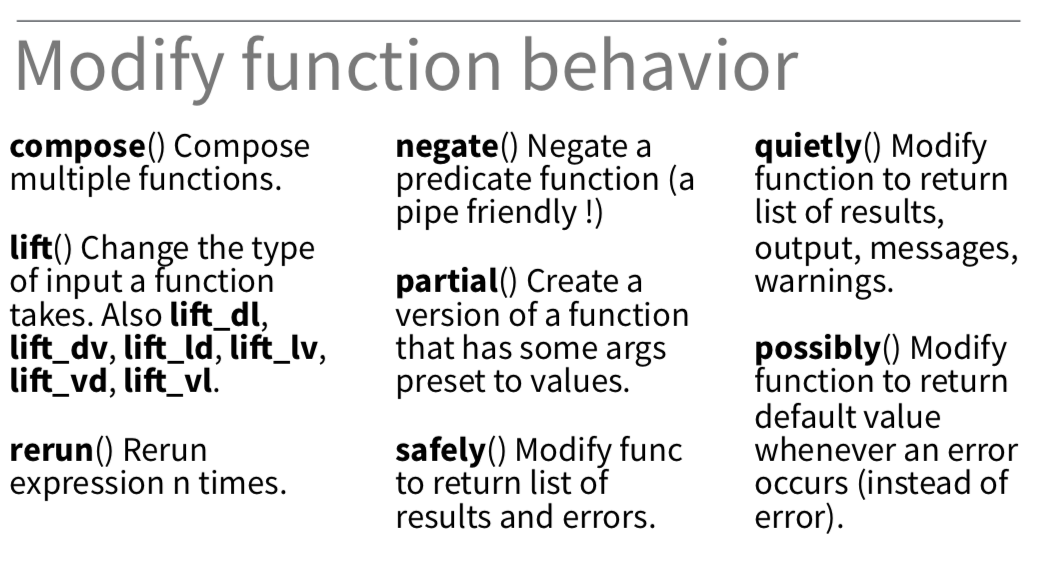
89 / 90
Learn more!
Jenny Bryan's purrr tutorial: A detailed introduction to purrr. Free online.
R for Data Science: A comprehensive but friendly introduction to the tidyverse. Free online.
RStudio Primers: Free interactive courses in the Tidyverse
90 / 90