Best Practices in R
2020-08-22
developed by Emil Hvitfeldt
Welcome!

Change Settings
Keyboard shortcut to open settings⌘ + , in Mac OS,ctrl + , in Windows
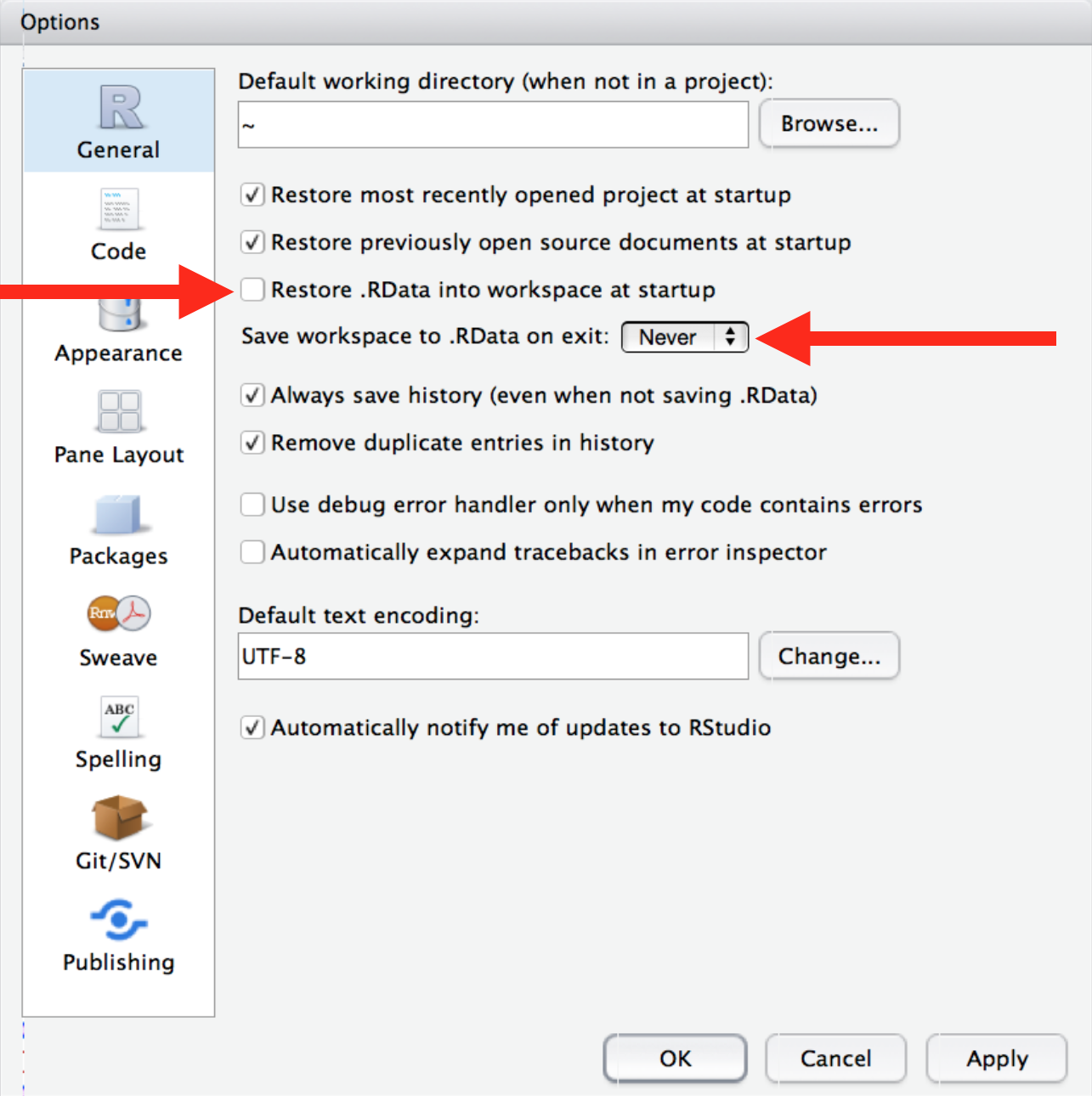
✓ - Uncheck "Restore .RData into work space at start up"
✓ - Set "Save work space to .Rdata on exit" to "Never"
Change Appearance
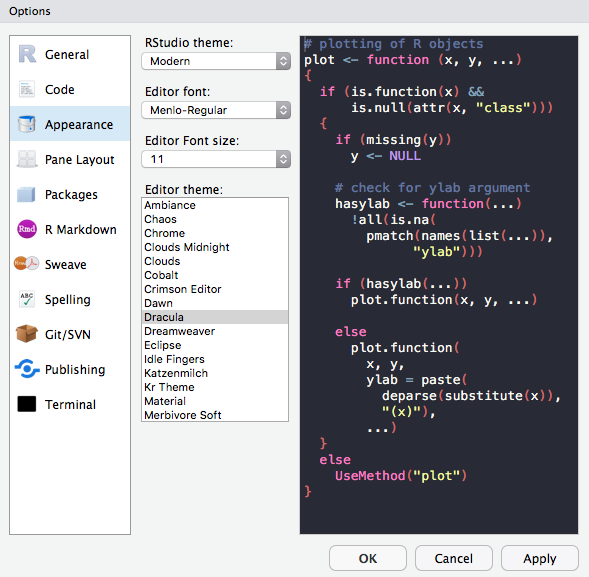
RStudio themes
Fonts
Font Sizes
Editor Themes
Pane layouts
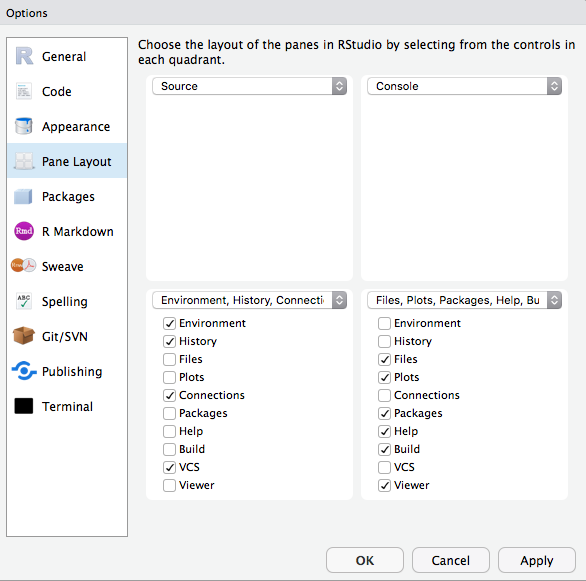
Change the layout of the panes
Source on top?
Source down to the right?
It's all up to you!
Pane layouts
Some like having both source and console open
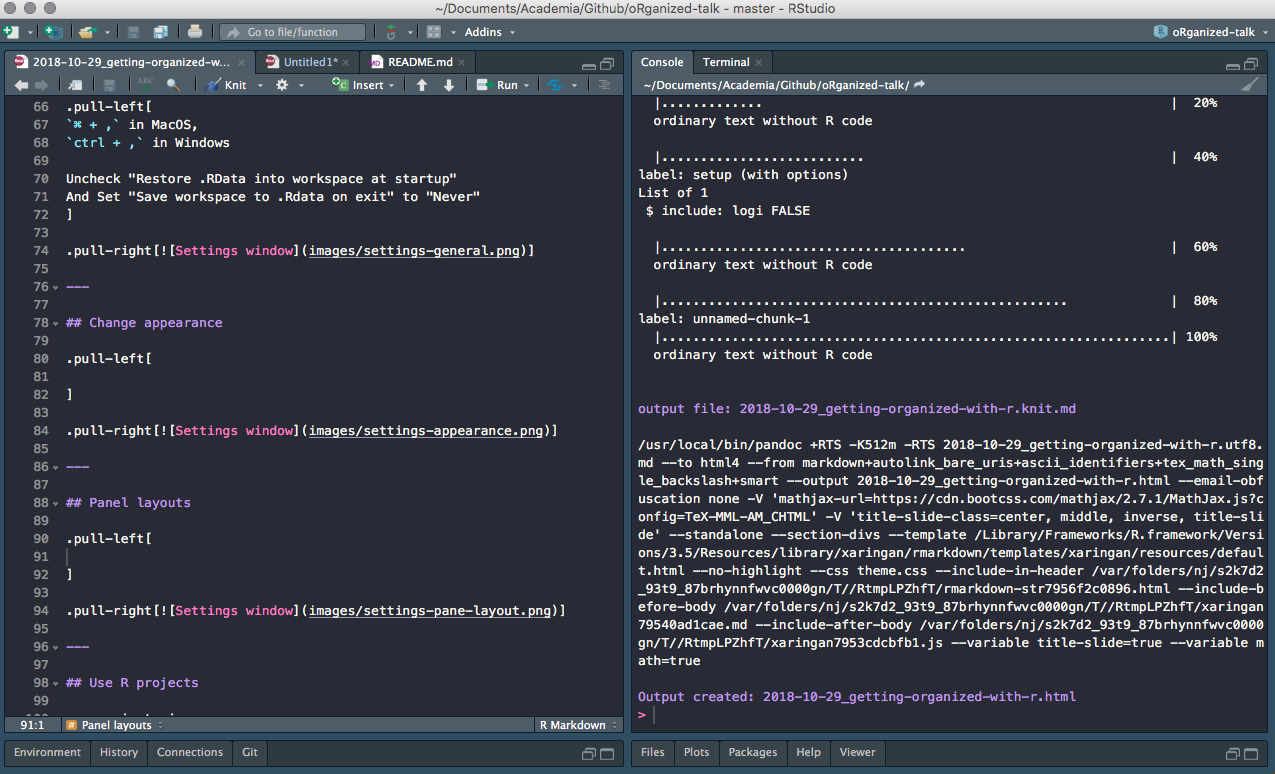
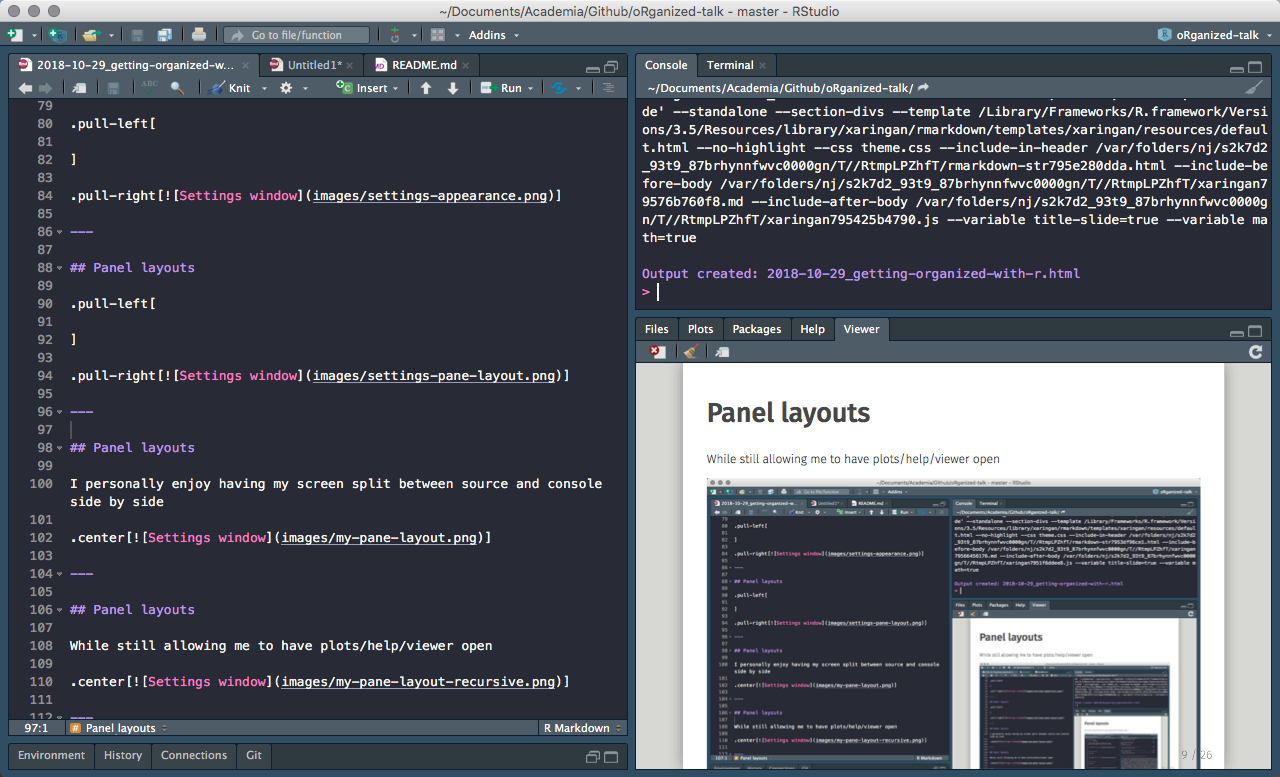
Pane layouts
...while still allowing to have viewer open
RStudio Projects
Keep all files from one project together. Use RStudio projects.
RStudio Projects
Keep all files from one project together. Use RStudio projects.
Self contained
RStudio Projects
Keep all files from one project together. Use RStudio projects.
Self contained
Project orientated
keep all the files associated with a project together — input data, R scripts, analytic results, figures.
usethis
usethis::create_project("project_name")
RStudio Projects - Creation 1 / 4
Click File > New Project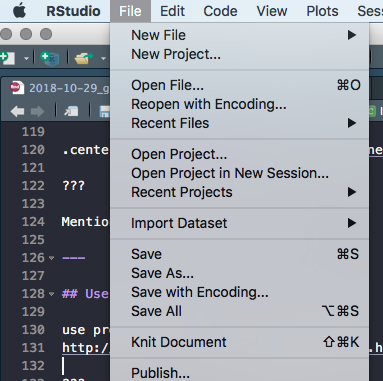
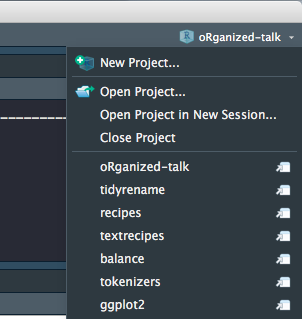
Or click on the upper right
RStudio Projects - Creation 2 / 4
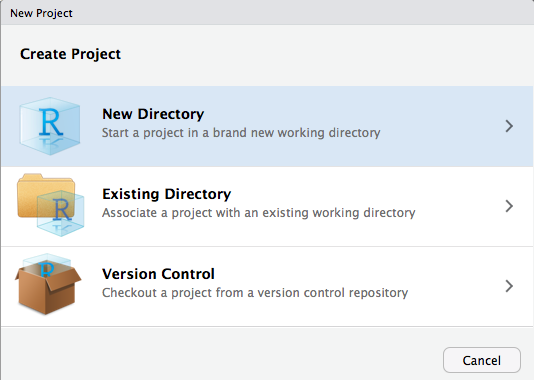
RStudio Projects - Creation 3 / 4
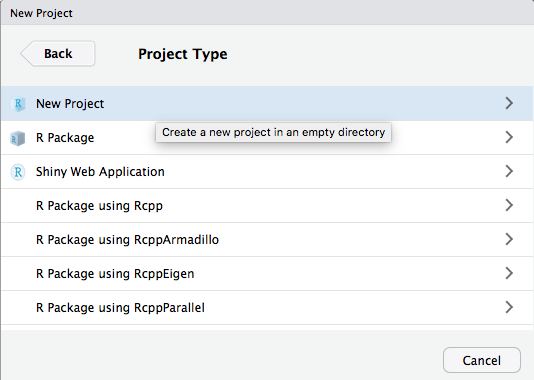
RStudio Projects - Creation 4 / 4
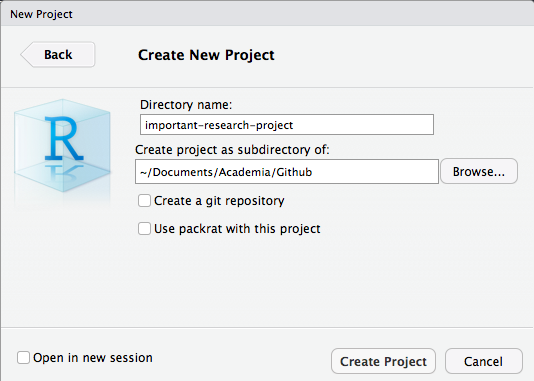
Folder Structure
Folder Structure
name_of_project|--raw_data |--WhateverData.xlsx |--report_2017.csv|--output_data |--summary2017.csv|--rmd |--01-analysis.Rmd|--docs |--01-analysis.html |--01-analysis.pdf|--scripts |--exploratory_analysis.R |--pdf_scraper.R|--figures |--weather_2017.png|--name_of_project.Rproj|--run_all.R- Raw data separate from cleaned data
- Reports and scrips are separated
- Generated and imported figures has its own place
- Numbered using 2 digits
- Reusable and easily understandable
Folder Structure
library(fs)folder_names <- c("raw_data", "output_data", "rmd", "docs", "scripts", "figures")dir_create(fldr_names)never modify raw data, only read (forever untouched)
Paths
library(tidyverse)# data importdata <- read_csv("/Users/Emil/Research/Health/amazing_data.csv")Paths
library(tidyverse)# data importdata <- read_csv("/Users/Emil/Research/Health/amazing_data.csv")## Error: '/Users/Emil/Research/Health/amazing_data.csv' does not exist.Paths
library(tidyverse)# data importdata <- read_csv("/Users/Emil/Research/Health/amazing_data.csv")## Error: '/Users/Emil/Research/Health/amazing_data.csv' does not exist.Only use relative paths, never absolute paths
Introducing the here package.
library(here)here()## [1] "/Users/Emil/Research/Health"library(here)data <- read_csv(here("raw_data", "amazing_data.csv"))Naming Things
Naming Things

- Organization
- Ease of use
There will be multi slides about naming
Naming Things - Files
NO
report.pdfreportv2.pdfreportthisisthelastone.pagesFigure 2.png 3465-234szx.rfoo.RYES
2018-10-01_01_report-for-cdc.pdf01_data.rmd01_data.pdf02_data-filtering.rmd02_data-filtering.pdfFollow narrative from folder structure slide
jenny Bryan naming things
- Avoid spaces, punctuation, special characters and case sensitivity
- Deliberate use of delimiters
- Describe the contents of the file
- Put something numeric first
- Left pad numbers with zeroes
- Use a standard date (YYYY-MM-DD)
to preserve chronological and logical ordering.
Naming Things - Files
library(fs)dir_ls("data/", regexp = "health-study")## 2018-02-23_health-study_power-100_group-A1.csv## 2018-02-23_health-study_power-100_group-B1.csv## 2018-02-23_health-study_power-100_group-C1.csv## 2018-02-23_health-study_power-200_group-A1.csv## 2018-02-23_health-study_power-200_group-B1.csv## 2018-02-23_health-study_power-200_group-C1.csvNaming Things - Files
library(fs)dir_ls("data/", regexp = "health-study")## 2018-02-23_health-study_power-100_group-A1.csv## 2018-02-23_health-study_power-100_group-B1.csv## 2018-02-23_health-study_power-100_group-C1.csv## 2018-02-23_health-study_power-200_group-A1.csv## 2018-02-23_health-study_power-200_group-B1.csv## 2018-02-23_health-study_power-200_group-C1.csvstringr::str_split_fixed(x, "[_\\.]", 5)## [,1] [,2] [,3] [,4] [,5] ## [1,] "2018-02-23" "health-study" "power-100" "group-A1" "csv"## [2,] "2018-02-23" "health-study" "power-100" "group-B1" "csv"## [3,] "2018-02-23" "health-study" "power-100" "group-C1" "csv"## [4,] "2018-02-23" "health-study" "power-200" "group-A1" "csv"## [5,] "2018-02-23" "health-study" "power-200" "group-B1" "csv"## [6,] "2018-02-23" "health-study" "power-200" "group-C1" "csv"- Avoid spaces, punctuation, special characters and case sensitivity
- Deliberate use of delimiters
- File name should describe the contents of the file
- Put something numeric first
- Left pad numbers with zeroes
- Use ISO 8601 standard for dates (YYYY-MM-DD)
Naming Things - Files
library(tidyverse)map_df(dir_ls("data/", regexp = "health-study"), read_csv)# ordir_ls("data/", regexp = "health-study") %>% map_df(read_csv)- Avoid spaces, punctuation, special characters and case sensitivity
- Deliberate use of delimiters
- File name should describe the contents of the file
- Put something numeric first
- Left pad numbers with zeroes
- Use ISO 8601 standard for dates (YYYY-MM-DD)
Naming Things - Objects
- Only use lowercase letters, numbers, and
_ - Use names that are not jargony, weight instead of K
- Use informative names
Naming Things - Objects
# BaddfetuningVar# Goodhealth_dataerrortuning_varlowercase letters + numbers = alpha-numeric characters (ish)
What To Avoid - attach()
Never use attach()
What To Avoid - attach()
Never use attach()
attach(mtcars)mean(mpg)## [1] 20.09062Loads lots of names into the search path, ambiguous selections.
What To Avoid - attach()
Never use attach()
attach(mtcars)mean(mpg)## [1] 20.09062Loads lots of names into the search path, ambiguous selections.
Try with() or withr instead
What To Avoid - attach()
Never use rm(list=ls())
What To Avoid - attach()
Never use rm(list=ls())
Instead, restart the R session
CTRL+SHIFT+F10 for Windows
CMD+SHIFT+ALT+F10 for Mac OS
R Markdown documents versus R scripts
You can use R scripts for simple self contained tasks.
source() R scripts into your R Markdown document where you will do analyses, visualizations and reporting.
R Markdown
- 01-import.R- 02-clean-names.R- 03-tidy.R- etcR Markdown
- 01-import.R- 02-clean-names.R- 03-tidy.R- etcInclude at the start of R Markdown file
{r load_scripts, include = FALSE}library(here)source(here("scripts", "01-import.R"))source(here("scripts", "02-clean-names.R"))source(here("scripts", "03-tidy.R"))Naming Chunks
Names can be placed after the comma
```{r, chunk-label, results='hide', fig.height=4}or before
```{r chunk-label, results='hide', fig.height=4}In general it is recommended to use alphabetic characters with words separated by - and avoid other characters. - Yihui Xie
- Makes navigating the R Markdown document easier
- Makes your R Markdown easier to understand
- Clarifies error reports or progress of knitting
- Caching when moving chunks around
Lower left corner of Rstudio have menu where sections and chunks can be selected with.
Caching on unnamed chunks are based on numbering.
Setup Chunk
In a fresh R Markdown document you see this
```{r setup, include=FALSE}knitr::opts_chunk$set(echo = TRUE)Setup Chunk
In a fresh R Markdown document you see this
```{r setup, include=FALSE}knitr::opts_chunk$set(echo = TRUE)The setup chunk is run before another code - use to your advantage
Setting figure path
Setting figure path
```{r setup, include=FALSE}knitr::opts_chunk$set(fig.path = "figures/")highlight use of fig.path option
fig.path: ('figure/'; character) prefix to be used for figure filenames (fig.path and chunk labels are concatenated to make filenames)
Styling Code
Use consistent style when writing code
Styling Code
Use consistent style when writing code
http://style.tidyverse.org/
All about preferences but keep it consistent!!!
Give examples of styles to follow
Use the styler package to style your code for you
Keep .Rprofile Clean
Your computer contains a file called .Rprofile.
This file runs first in every session. Think of it as configuration file.
Keep .Rprofile Clean
Your computer contains a file called .Rprofile.
This file runs first in every session. Think of it as configuration file.
options(stringsAsFactors = FALSE)options(max.print = 100)Keep .Rprofile Clean
Only put interactive code in
Yes
# add this with usethis::use_usethis()library(usethis)No
library(tidyverse)Use it to change options and load packages
Comment Your Code
Functions: Arguments and purpose
Code: What or why, NOT how
Comment Your Code
Functions: Arguments and purpose
Code: What or why, NOT how
# Takes a data.frame (data) and replaces the columns with the names# (names) and converts them from factor variable to character # variables. Keeps characters variables unchanged.factor_to_text <- function(data, names) { for (i in seq_along(names)) { if(is.factor(data[, names[i], drop = TRUE])) data[, names[i]] <- as.character.factor(data[, names[i], drop = TRUE]) } data}Updating R and RStudio
The most recent version of R can be downloaded from The Comprehensive R Archive Network (CRAN)
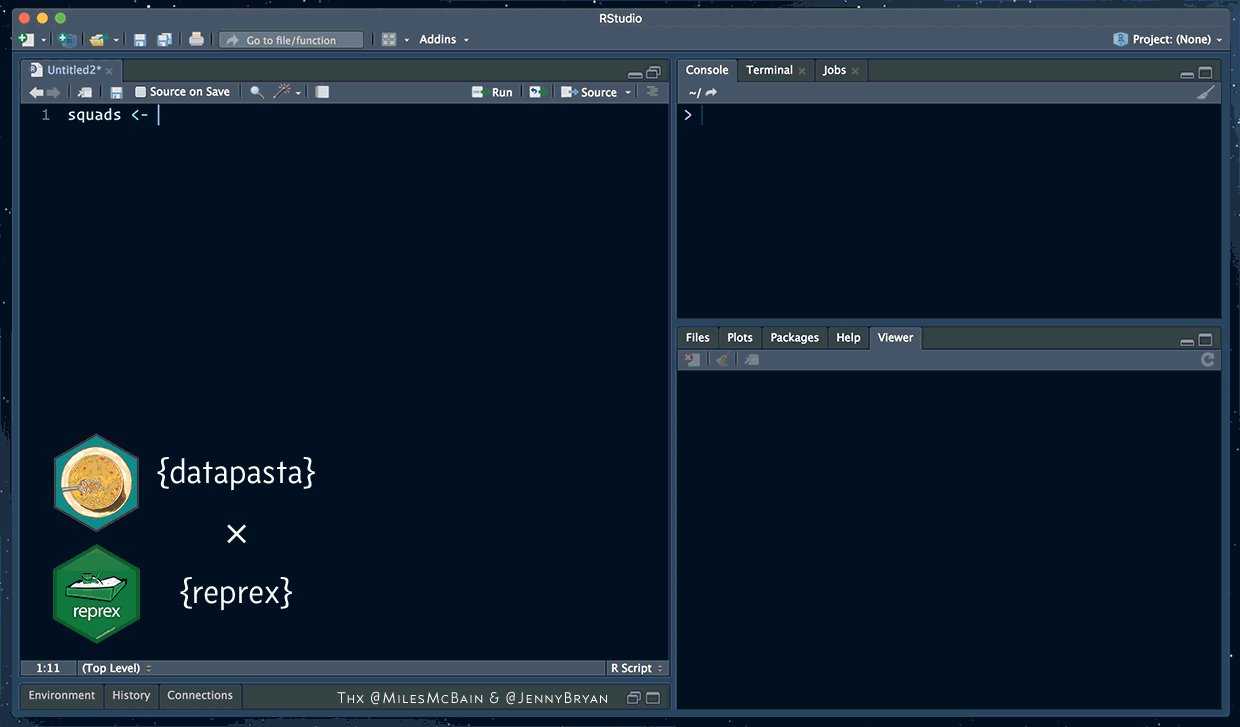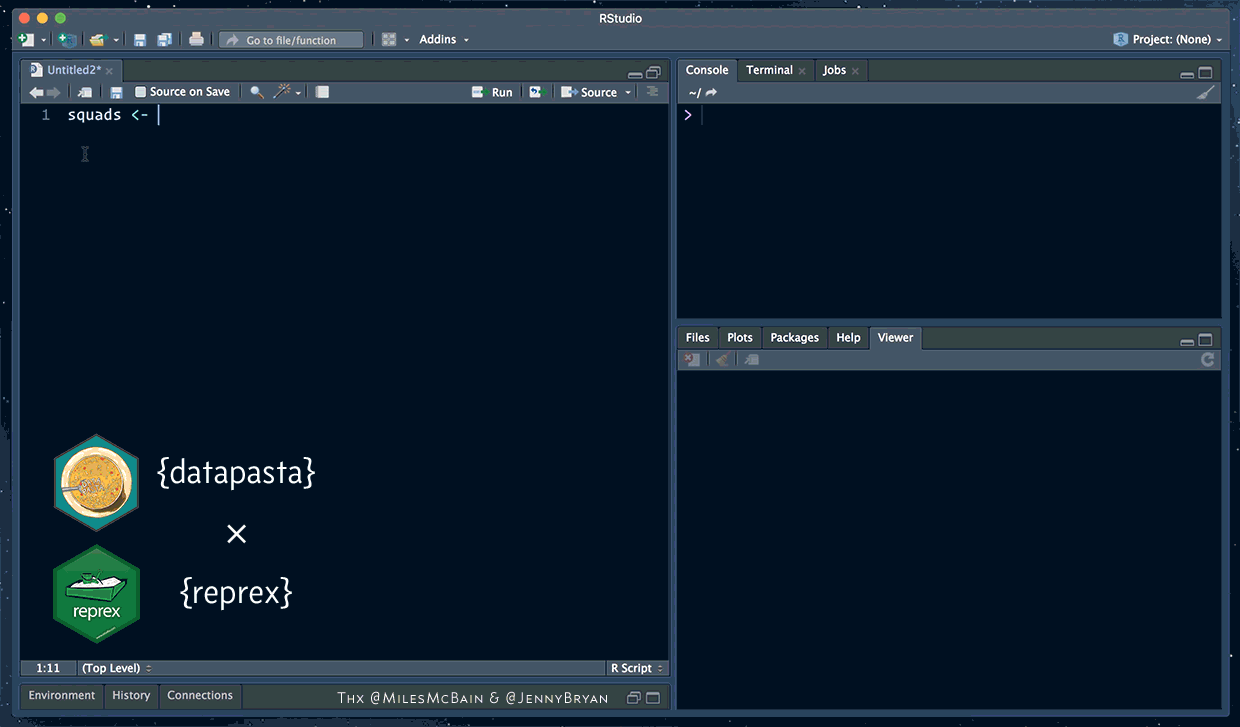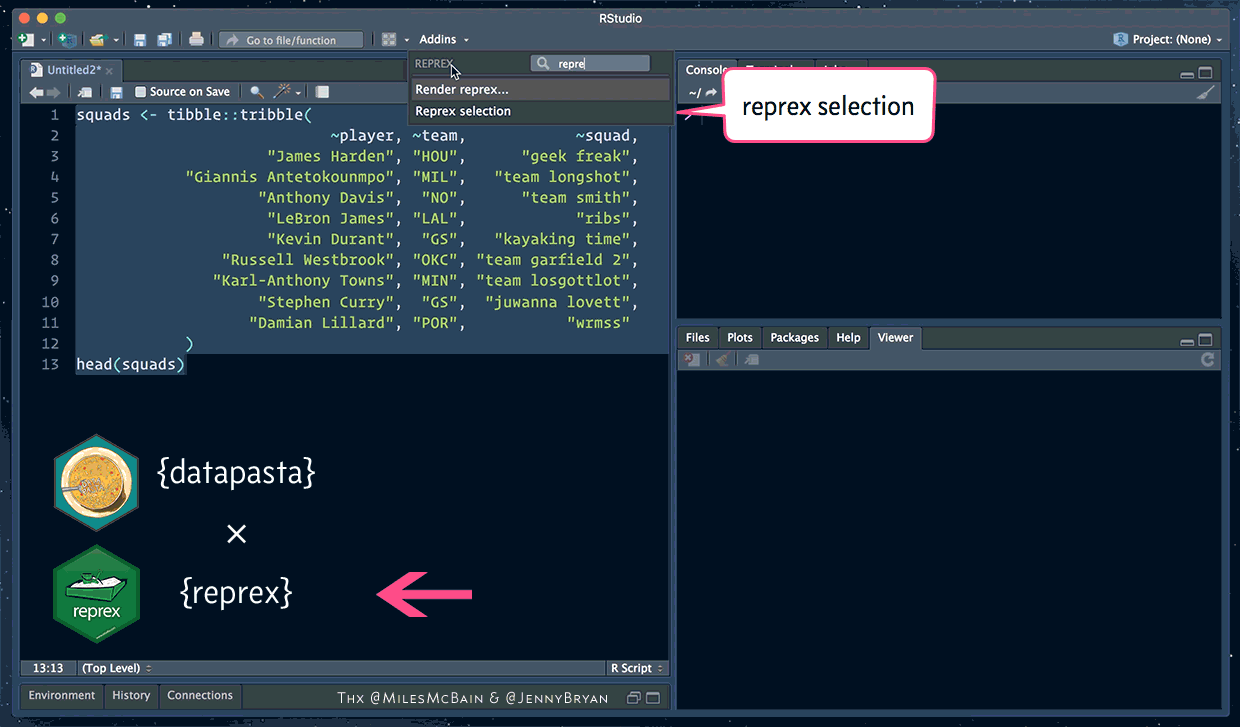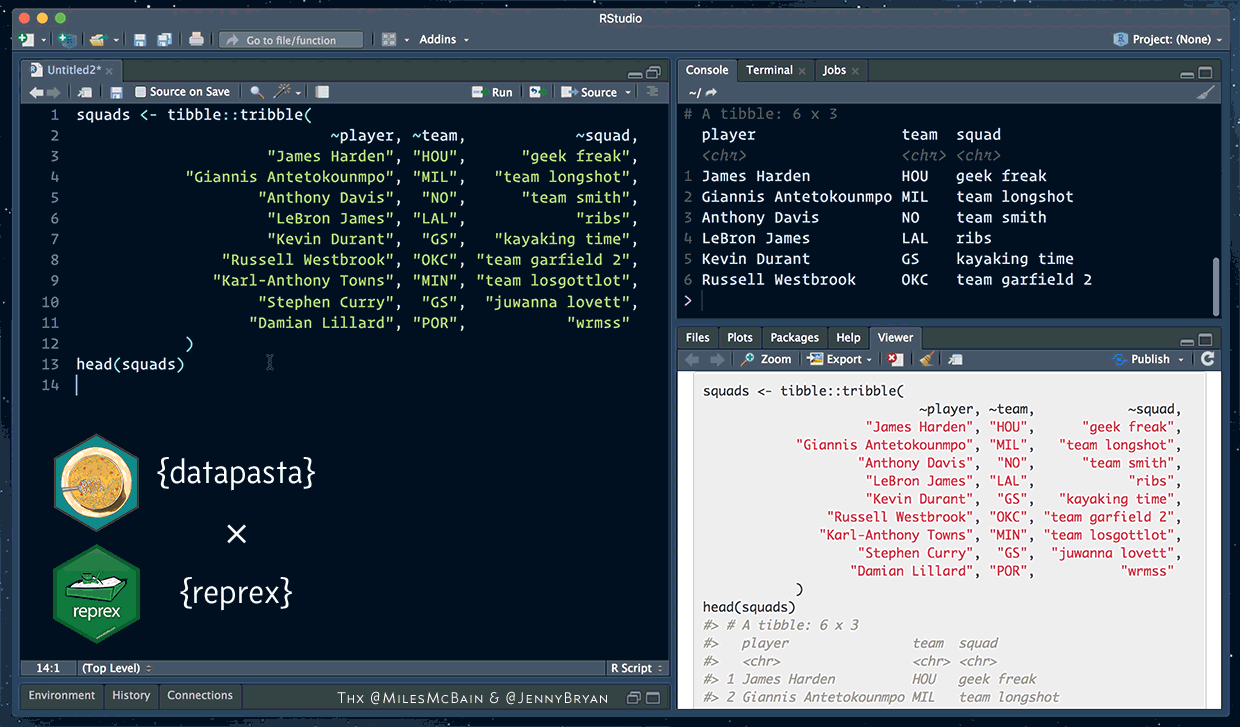
How to ask for help (datapasta and reprex)
The reprex package helps you create a reproducible example
datapasta lets you easy copy + paste small samples of data into RStudio
How to ask for help (reprex)
Check out the package website and RStudio webinar on creating reproducible examples
 Art by Allison Horst
Art by Allison Horst
Where to get help
RStudio has a helpful community if you have questions (everyone does!)
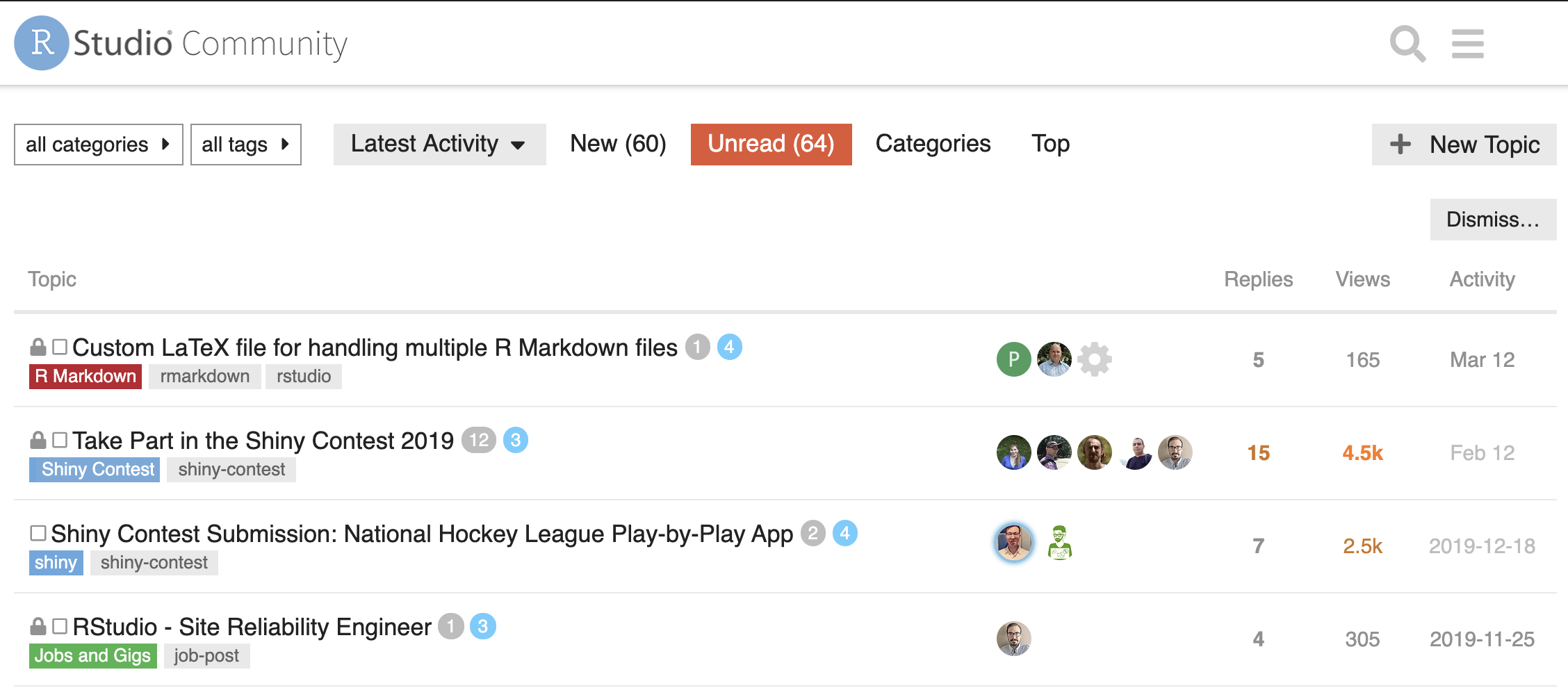
RStudio Community:
RStudio has a dedicated forum for questions related to R and RStudio: https://community.rstudio.com/
Where else to get help
Stack Overflow
Check out the questions tagged r on Stack Overflow: https://stackoverflow.com/questions/tagged/r
#rstats on Twitter
If you have a Twitter account, check out #rstats: https://twitter.com/hashtag/rstats

Art by Allison Horst